In this post we’ll describe how we’ve setup Atlas to run within a Google Cloud environment, using BigQuery as it’s source database.
Why Atlas?
But why setup Atlas anyway? Atlas is a very neat tool to conduct efficient analyses on standardized observational data converted to the OMOP Common Data Model (CDM). By converting healthcare data to a standardized format, researchers can easily apply their analyses to multiple data sources. Atlas facilitates these analyses and can be used out of the box to gain quick insights into OMOP CDM data. Also, since it is open source software, it is freely available and has a community of users and developers to help you on your way. Atlas mainly evolves around concept sets and cohorts which researches can define based on the standardized vocabularies. Using these concept sets and cohorts, you can create groups of patients based on for example an exposure to a drug or diagnosis of a condition.
Although this might sound very easy, it takes quite some steps to get Atlas up and running. We chose to setup Atlas in our Google Cloud Platform (GCP) environment. Of course you can use any other cloud platform, or setup Atlas on premises. However, by showing it is possible to setup Atlas on a cloud platform such as GCP, we hope to prove you don’t need a large organization with existing IT infrastructure to start with data standardization in healthcare.
In this blog we’ll look at:
- Database setup (BigQuery)
- Creating a virtual machine to host atlas
- Network and firewall rules
- Using Docker and Docker Compose
- Installing database drivers
- Refreshing your OMOP CDM data
- Debugging
Prerequisites
Since we’re running this project on Google Cloud, a Google Cloud project would come in handy. Also, this post assumes you already have data available in the OMOP CDM format.
Prerequisites:
- Google Cloud project
- BigQuery dataset containing OMOP data
Creating the BigQuery tables
To run Atlas, you need to have a database containing data formatted according to the OMOP Common Data Model. Usually you have an ETL-process in place to do just that. We’ve for instance designed an ETL in R to convert GemsTracker data from a MySQL database into OMOP CDM.
Once you have an ETL in place, you need a database to store the resulting data. Since we’re running this project in Google Cloud, we’ve opted for BigQuery. Luckily OHDSI provides us with the (almost) complete queries to initiate the necessary BigQuery tables. First, create a new BigQuery dataset dedicated to this data, and then run the BigQuery DDL. Simply replace @cdmDatabaseSchema with the name of the dataset you created, and you’re good to go. For now I’ll call this dataset cdm, but you can pick anything you like. When we created our tables, there were still some tiny bugs in the DDL code, which you’ll spot soon enough. Also, you can simply use the version we prepared.
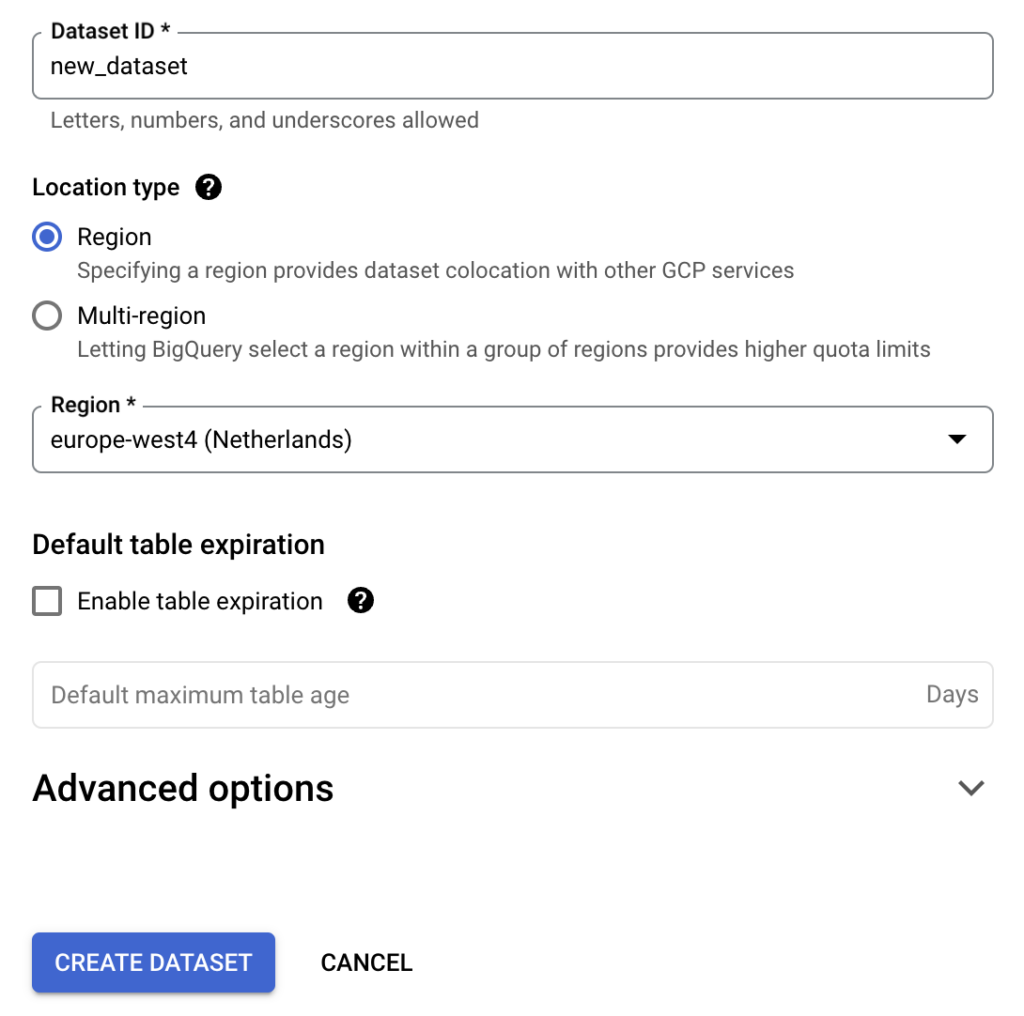
Lastly, we’ll also create two empty datasets for temporary data and results data, we’ll call these temp and results. These datasets are needed later in the process.
Creating a VM to host Atlas
Next, we’ll need a virtual machine to host Atlas. For this we’ll use Google Compute Engine. To make sure our VM is secure, we’ll use a private VPC and a VM without a public IP.
VPC network
First we need to create a private VPC-network. When creating a VPC-network, Google will also ask you to create a new subnet. You can do this via the Google interface, or run it via cli, using the code below. Of course, you can modify the names and region.
gcloud compute networks create new-vpc-network --project=new-project --subnet-mode=custom --mtu=1460 --bgp-routing-mode=regional && gcloud compute networks subnets create new-subnet --project=new-project --range=10.0.0.0/24 --stack-type=IPV4_ONLY --network=new-vpc-network --region=europe-west4Firewall rules
Next, we’re going to create a new firewall rule. This firewall will allow us to use Identity-Aware Proxy (IAP) TCP forwarding to connect to the VM without the need of a public IP. You can check out the Google documentation for some more information about IAP.
We’ll create a firewall rule to allow ingress from IAP via port 22 (SSH). We’re going to limit this access to IP range 35.235.240.0/20, which corresponds to the IP’s Google uses internally. Also, we’ll attach a network tag allow-ingress-from-iap to the firewall rule. This enables us to apply this rule only to VM’s containing the same network tag.
gcloud compute --project=new-project firewall-rules create allow-ingress-from-iap --direction=INGRESS --priority=1000 --network=new-vpc-network --action=ALLOW --rules=tcp:22 --source-ranges=35.235.240.0/20 --target-tags=allow-ingress-from-iapCompute instance
This is where we’ll create the VM to host Atlas. You can pick any machine you like, but we’re going to use a basic Ubuntu 22 image. Make sure your boot disk isn’t too small, and don’t forget to disable the public IP.
gcloud compute instances create atlas-instance --project=new-project --zone=europe-west4-a --machine-type=e2-medium --network-interface=stack-type=IPV4_ONLY,subnet=new-subnet,no-address --maintenance-policy=MIGRATE --provisioning-model=STANDARD --service-account=xyz-compute@developer.gserviceaccount.com --scopes=https://www.googleapis.com/auth/devstorage.read_only,https://www.googleapis.com/auth/logging.write,https://www.googleapis.com/auth/monitoring.write,https://www.googleapis.com/auth/servicecontrol,https://www.googleapis.com/auth/service.management.readonly,https://www.googleapis.com/auth/trace.append --tags=allow-ingress-from-iap,http-server,https-server --create-disk=auto-delete=yes,boot=yes,device-name=atlas-instance,image=projects/ubuntu-os-cloud/global/images/ubuntu-2204-jammy-v20230429,mode=rw,size=50,type=projects/new-project/zones/europe-west4-a/diskTypes/pd-balanced --no-shielded-secure-boot --shielded-vtpm --shielded-integrity-monitoring --labels=ec-src=vm_add-gcloud --reservation-affinity=anyConfiguring the VM
Installing Docker
Now we have our VM up and running, it’s time to prepare it to run Atlas. Since we’re using Broadsea, the first thing we need is Docker and Docker Compose. Luckily, DigitalOcean already prepared a perfect tutorial of how to install Docker on Ubuntu. These next steps are taken from their guide.
sudo apt update
sudo apt install apt-transport-https ca-certificates curl software-properties-common
curl -fsSL https://download.docker.com/linux/ubuntu/gpg | sudo gpg --dearmor -o /usr/share/keyrings/docker-archive-keyring.gpg
echo "deb [arch=$(dpkg --print-architecture) signed-by=/usr/share/keyrings/docker-archive-keyring.gpg] https://download.docker.com/linux/ubuntu $(lsb_release -cs) stable" | sudo tee /etc/apt/sources.list.d/docker.list > /dev/null
sudo apt update
apt-cache policy docker-ce
sudo apt install docker-ce
sudo systemctl status docker
sudo apt install docker-composeClone Broadsea repository
To run Broadsea, we need to get the Broadsea repository to our VM. Make sure your Git account is authenticated and clone the Broadsea repo. For this tutorial, we’re going to use the most recent version (v3.0.0).
git clone git@github.com:OHDSI/Broadsea.git --branch v3.0.0
git clone git@github.com:OHDSI/WebAPI.git --branch v2.13.0Service account
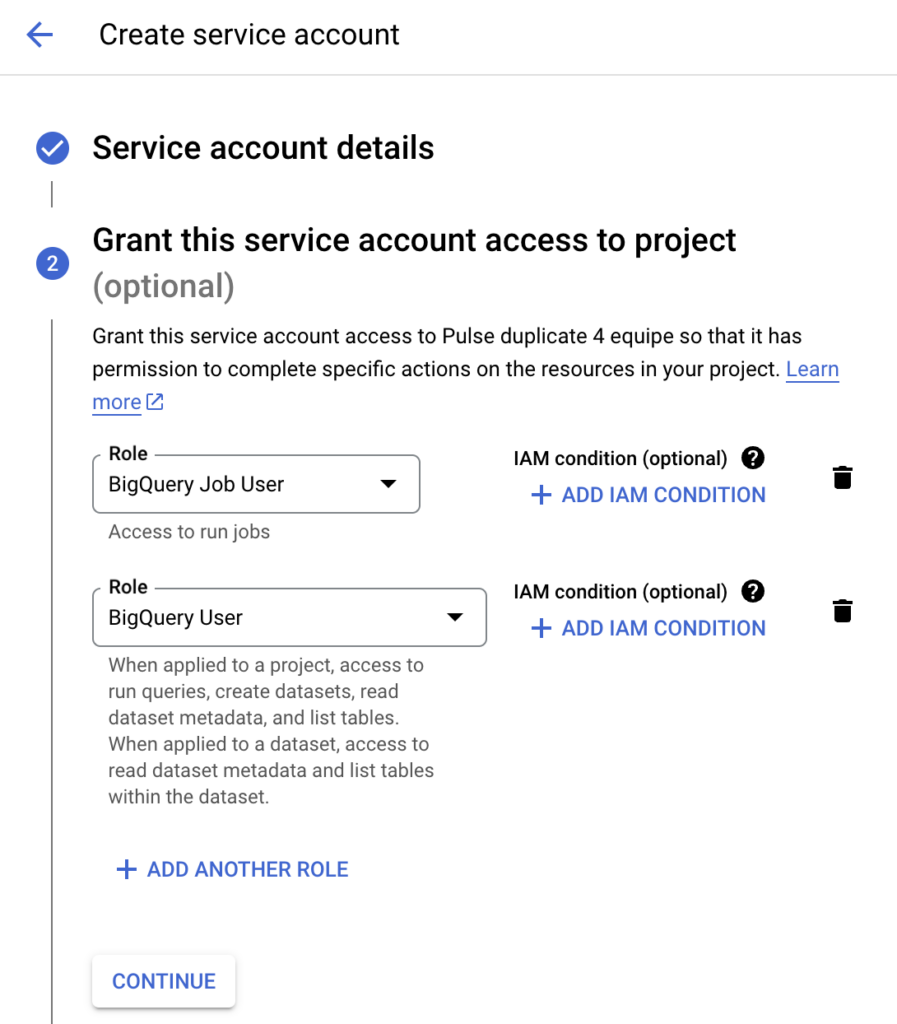
Also, we need a Google service account with the correct rights to run BigQuery jobs. Create a new service account using Google IAM. Grant this service account at least the BigQuery User and BigQuery Job User roles, and download the service account key. We need to upload this service account key, which is a json file, to the VM. If you SSH into the VM via the Google Compute Engine page, Google provides you with a nice upload button, which makes uploading file very easy.
BigQuery drivers
Broadsea comes with PostgreSQL drivers included. However, to use BigQuery, we need to download the appropriate drivers first, and save them in the Broadsea folder.
sudo apt-get install unzip
sudo curl -o SimbaJDBCDriverforGoogleBigQuery42_1.2.25.1029.zip https://storage.googleapis.com/simba-bq-release/jdbc/SimbaJDBCDriverforGoogleBigQuery42_1.2.25.1029.zip
sudo unzip SimbaJDBCDriverforGoogleBigQuery42_1.2.25.1029.zip -d bigquery
sudo rm SimbaJDBCDriverforGoogleBigQuery42_1.2.25.1029.zipInstalling WebAPI
Because we’re using our own WebAPI drivers, we’ll need to do a custom WebAPI installation. First we’ll copy the downloaded BigQuery drivers into the cloned WebAPI folder.
sudo cp -R bigquery WebAPINext, we’ll add the following line to the Dockerfile, to make sure the BigQuery drivers are available during installation. For this you can any text editor you like, e.g. nano: sudo nano Dockerfile.
COPY /bigquery/ /code/src/main/extras/bigquery/As described in the WebAPI documentation, we also need to create our own settings.xml file. For now it’s sufficient to copy the sample_settings.xml into a new folder.
sudo mkdir WebAPIConfig
sudo cp sample_settings.xml WebAPIConfig/settings.xmlAlso, the WebAPI contains a pom.xml file with different profiles for each possible data source. We’re going to use the webapi-bigquery profile. But since we downloaded our own drivers, we need to modify the profile to match the version numbers of the downloaded drivers. The following piece of xml should match the correct version numbers. You can copy and paste this into your pom.xml file.
<profile>
<id>webapi-bigquery</id>
<properties>
<bigquery.enabled>true</bigquery.enabled>
<!-- BigQuery JDBC driver path -->
<bigquery.classpath>${basedir}/src/main/extras/bigquery</bigquery.classpath>
</properties>
<dependencies>
<dependency>
<groupId>com.google.api-client</groupId>
<artifactId>google-api-client</artifactId>
<version>2.1.1</version>
</dependency>
<dependency>
<groupId>com.google.apis</groupId>
<artifactId>google-api-services-bigquery</artifactId>
<version>v2-rev20221028-2.0.0</version>
</dependency>
<dependency>
<groupId>com.google.cloud</groupId>
<artifactId>google-cloud-bigquery</artifactId>
<version>2.9.4</version>
</dependency>
<dependency>
<groupId>com.google.http-client</groupId>
<artifactId>google-http-client</artifactId>
<version>1.42.3</version>
</dependency>
<dependency>
<groupId>com.google.oauth-client</groupId>
<artifactId>google-oauth-client</artifactId>
<version>1.34.1</version>
</dependency>
<dependency>
<groupId>com.google.cloud</groupId>
<artifactId>google-cloud-bigquerystorage</artifactId>
<version>2.26.0</version>
</dependency>
<dependency>
<groupId>com.google.auth</groupId>
<artifactId>google-auth-library-oauth2-http</artifactId>
<version>1.12.1</version>
</dependency>
<dependency>
<groupId>com.google.auth</groupId>
<artifactId>google-auth-library-credentials</artifactId>
<version>1.12.1</version>
</dependency>
<dependency>
<groupId>com.google.api</groupId>
<artifactId>gax</artifactId>
<version>2.19.5</version>
</dependency>
<dependency>
<groupId>com.google.api</groupId>
<artifactId>gax-grpc</artifactId>
<version>2.19.5</version>
</dependency>
<dependency>
<groupId>com.google.api.grpc</groupId>
<artifactId>proto-google-cloud-bigquerystorage-v1beta1</artifactId>
<version>0.150.0</version>
</dependency>
<dependency>
<groupId>io.opencensus</groupId>
<artifactId>opencensus-contrib-http-util</artifactId>
<version>0.31.1</version>
</dependency>
</dependencies>
<build>
<plugins>
<plugin>
<groupId>org.apache.maven.plugins</groupId>
<artifactId>maven-install-plugin</artifactId>
<version>2.5.2</version>
<executions>
<execution>
<id>google-api-client</id>
<phase>initialize</phase>
<goals>
<goal>install-file</goal>
</goals>
<configuration>
<groupId>com.google.api-client</groupId>
<artifactId>google-api-client</artifactId>
<version>2.1.1</version>
<packaging>jar</packaging>
<file>${bigquery.classpath}/google-api-client-2.1.1.jar</file>
</configuration>
</execution>
<execution>
<id>google-api-services-bigquery</id>
<phase>initialize</phase>
<goals>
<goal>install-file</goal>
</goals>
<configuration>
<groupId>com.google.apis</groupId>
<artifactId>google-api-services-bigquery</artifactId>
<version>v2-rev20221028-2.0.0</version>
<packaging>jar</packaging>
<file>${bigquery.classpath}/google-api-services-bigquery-v2-rev20221028-2.0.0.jar</file>
</configuration>
</execution>
<execution>
<id>google-cloud-bigquery</id>
<phase>initialize</phase>
<goals>
<goal>install-file</goal>
</goals>
<configuration>
<groupId>com.google.cloud</groupId>
<artifactId>google-cloud-bigquery</artifactId>
<version>2.9.4</version>
<packaging>jar</packaging>
<file>${bigquery.classpath}/GoogleBigQueryJDBC42.jar</file>
</configuration>
</execution>
<execution>
<id>google-http-client</id>
<phase>initialize</phase>
<goals>
<goal>install-file</goal>
</goals>
<configuration>
<groupId>com.google.http-client</groupId>
<artifactId>google-http-client</artifactId>
<version>1.42.3</version>
<packaging>jar</packaging>
<file>${bigquery.classpath}/google-http-client-1.42.3.jar</file>
</configuration>
</execution>
<execution>
<id>google-oauth-client</id>
<phase>initialize</phase>
<goals>
<goal>install-file</goal>
</goals>
<configuration>
<groupId>com.google.oauth-client</groupId>
<artifactId>google-oauth-client</artifactId>
<version>1.34.1</version>
<packaging>jar</packaging>
<file>${bigquery.classpath}/google-oauth-client-1.34.1.jar</file>
</configuration>
</execution>
<execution>
<id>google-cloud-bigquerystorage</id>
<phase>initialize</phase>
<goals>
<goal>install-file</goal>
</goals>
<configuration>
<groupId>com.google.cloud</groupId>
<artifactId>google-cloud-bigquerystorage</artifactId>
<version>2.26.0</version>
<packaging>jar</packaging>
<file>${bigquery.classpath}/google-cloud-bigquerystorage-2.26.0.jar</file>
</configuration>
</execution>
</executions>
</plugin>
</plugins>
</build>
</profile>And then we’re ready to build our WebAPI image, using the BigQuery profile, next to the default PostgreSQL profile.
sudo docker build -t broadsea-webapi-bigquery --build-arg MAVEN_PROFILE=webapi-postgresql,webapi-bigquery .Docker Compose
Now, our WebAPI image is ready to go. However, we still need the rest of the Broadsea ecosystem. We already cloned the Broadsea repository and downloaded the BigQuery drives, so lets start by copying those drivers to the correct folder.
sudo cp -R bigquery BroadseaNext to the drivers, we also uploaded a Google service account. Lets also move this to our Broadsea folder.
sudo cp service-account.json BroadseaNow we have the BigQuery drivers and service account available, we need to mount the to our Dockerfile in the docker-compose.yml. Add the following lines to the broadsea-hades service:
volumes:
- ./bigquery:/tmp/drivers/:ro
- ./service-account-key.json:/tmp/google/service-account-key.jsonIn Broadsea 3.0, the WebAPI has its own ohdsi-webapi.yml file, which is referred to in docker-compose.yml. Add the following lines to this file:
volumes:
- ./bigquery:/tmp/drivers/:ro
- ./service-account-key.json:/var/lib/ohdsi/webapi/service-account-key.jsonAlso, since we’re using a custom installation of the WebAPI, we need to mention this in the docker-compose.yml file, instead of simply retrieving the latest WebAPI version from Docker Hub. Therefore replace image: docker.io/ohdsi/webapi:latest with image: broadsea-webapi-bigquery, which is the name we gave to our WebAPI image earlier.
Lastly, the are loads of environment variables which are referred to by ${VARIABLE_NAME} in the docker-compose file. These variabeles are defined in the .env file, which is also were you can change them. For example, you can set the username and password of your Hades user.
Now everything is ready to go, lets start those containers!
sudo docker-compose --profile default up -dAccessing Atlas
When your services are up, Atlas is ready to use. Since we created our container in a private VPC network, we’ll need IAP (identity-aware proxy) to access it. Make sure your user has the correct rights to access the VM via IAP. These permissions can be granted for every VM separately. Just find IAP in Google Cloud, and use the IAP-secured Tunnel User role.
Now we are ready to create an IAP-tunnel to the http-port to which Atlas is exposed (80) and the port which we use locally (8001), since 80 is usually reserved.
gcloud compute start-iap-tunnel atlas-instance 80 --local-host-port=localhost:8001 --zone=europe-west4-aAdding a new data source
Now we have Atlas up and running, but we haven’t added our own data source yet. By default the Broadsea version of Atlas contains a demo dataset (Eunomia), but this isn’t what we’re looking for.
Results schema
Earlier in this post we mentionted the creation of a results dataset in BigQuery. However, we did not create any tables yet to hold the data. Now the WebAPI is active, we can use the following url to generate the queries to create the correct tables. Make sure your IAP-tunnel is active, and you check if the dataset names match those in your BigQuery environment.
http://127.0.0.1:8001/WebAPI/ddl/results?dialect=bigquery&schema=results&vocabSchema=cdm&tempSchema=temp&initConceptHierarchy=trueThe resulting queries can be ran in BigQuery.
Insert queries
To add a data source, we need to tell the WebAPI where to find it. This can be done by adding new records to the webapi.source and the webapi.source_daimon tables, which reside in the broadsea-atlasdb container. How you can add records to these tables is also described in the WebAPI Wiki, but we’ll give an example for BigQuery.
First, we need to enter the broadsea-atlasdb container.
sudo docker exec -it broadsea-atlasdb /bin/bashIf we’re in, we can enter the internal PostgreSQL database. This is just a database for internal use, and differs from the BigQuery database which hosts your actual OMOP data.
psql --host localhost --username postgresWe’ll start by adding a new record to webapi.source. This contains the name and id of your data source, but most importantly a connection string for BigQuery. Make sure to check this connection string, and adjust the name of the GCP-project and service account to match your own set-up.
-- OHDSI CDM source
INSERT INTO webapi.source( source_id, source_name, source_key, source_connection, source_dialect, is_cache_enabled)
VALUES (2, 'NewCDM', 'cdm',
'jdbc:bigquery://https://www.googleapis.com/bigquery/v2:443;ProjectId=new-project;OAuthType=0;OAuthServiceAcctEmail=service-account@new-project.iam.gserviceaccount.com;OAuthPvtKeyPath=/var/lib/ohdsi/webapi/service-account-key.json;', 'bigquery', 't');Next, we’ll add four records to webapi.source_daimon table. These contain the different BigQuery datasets which we mentioned at the beginning of this post. These datasets will all contain different types of data needed to run Atlas. Make sure the source_id in both tables is the same.
-- CDM daimon
INSERT INTO webapi.source_daimon( source_daimon_id, source_id, daimon_type, table_qualifier, priority) VALUES (4, 2, 0, 'cdm', 0);
-- VOCABULARY daimon
INSERT INTO webapi.source_daimon( source_daimon_id, source_id, daimon_type, table_qualifier, priority) VALUES (5, 2, 1, 'cdm', 10);
-- RESULTS daimon
INSERT INTO webapi.source_daimon( source_daimon_id, source_id, daimon_type, table_qualifier, priority) VALUES (6, 2, 2, 'results', 0);
-- TEMP daimon
INSERT INTO webapi.source_daimon( source_daimon_id, source_id, daimon_type, table_qualifier, priority) VALUES (7, 2, 5, 'temp', 0);Finally, let’s restart the WebAPI, so the changes can take effect.
sudo docker restart ohdsi-webapiUpdate record counts
When you open Atlas again, you should now see your own data source available under data sources. However, if you’ll search for several concept_id’s present in you data, you’ll notice that most of them are still empty. Refreshing the record counts is done in a view steps, which we’ll discuss below.
Achilles
Most of the work is done by Achilles, which is an R package developed by OHDSI. To use the Achilles packages, we first need to establish a connection to BigQuery, using the DatabaseConnector package.
connection_details <- DatabaseConnector::createConnectionDetails(
dbms = "bigquery",
connectionString = "jdbc:bigquery://https://www.googleapis.com/bigquery/v2:443;ProjectId=new-project;OAuthType=0;OAuthServiceAcctEmail=service-account@new-project.iam.gserviceaccount.com;OAuthPvtKeyPath=/var/lib/ohdsi/webapi/service-account-key.json;Timeout=1000",
user = "",
password = "",
pathToDriver = "/tmp/drivers/bigquqery/"
)
connection <- DatabaseConnector::connect(connection_details)When the database connection is established, we can update all relevant tables using the achilles() function. This function can be executed directly on the database, or ran in sqlOnly mode, which returns and set of queries to run manually. At the moment of writing however, the Achilles packages doesn’t seem to be fully developed for BigQuery, and still has some syntactical issues. Therefore, we’re going to use the sqlOnly mode, and make some manual adjustments afterwards.
Achilles::achilles(
connectionDetails = connection_details,
cdmDatabaseSchema = "cdm",
resultsDatabaseSchema = "results",
scratchDatabaseSchema = "temp",
vocabDatabaseSchema = "cdm",
sourceName = "newCDM",
createTable = FALSE,
defaultAnalysesOnly = FALSE,
dropScratchTables = TRUE,
sqlOnly = TRUE,
sqlDialect = "bigquery",
optimizeAtlasCache = TRUE,
cdmVersion = "5.4"
)By default, the achilles() function now saves an achilles.sql file in the output folder of your current working directory. If you read this query into an R object, you can do some fixed programatically, which will save you a lot of time. We’ve used something like here, but this can differ based on your data and Achilles version.
library(readr)
library(stringr)
query <- readr::read_file("output/achilles.sql")
# Add drop table statements
create_temp_table <- str_extract_all(query, "CREATE TABLE temp.tmpach_[a-z0-9_]+")[[1]]
create_results_table <- str_extract_all(query, "CREATE TABLE results.xrhqxw6s[a-z0-9_]+")[[1]]
create_temp_table <- c(create_temp_table, create_results_table)
for (create in create_temp_table) {
table_name <- gsub("CREATE TABLE ", "", create)
query <- gsub(
paste0(create, "[^0-9]"),
paste0("DROP TABLE IF EXISTS ", table_name, "; ", create),
query
)
}
# Fix incorrect data type
query <- gsub("numeric\\(7,6\\)", "numeric", query)
# Fix too complex query
tmpach_2004 <- paste0(
"CREATE TABLE temp.tmpach_2004",
gsub(";.*", "", gsub(".*CREATE TABLE temp.tmpach_2004", "", query)),
";"
)
tmpach_2004 <- gsub("WITH rawdata as \\(", "", tmpach_2004)
tmpach_2004 <- gsub("SELECT \\* FROM rawdata", "", tmpach_2004)
tmpach_2004 <- gsub(" union all", "; insert into temp.tmpach_2004", tmpach_2004)
query <- paste0(
gsub("CREATE TABLE temp.tmpach_2004.*", "", query),
tmpach_2004,
"-- 210",
gsub(".*\\-\\- 210", "", query)
)
# Fix intersect
query <- gsub(" intersect", " intersect distinct", query)
write_file(query, "achilles_new.sql")Now run the resulting queries via the BigQuery interface. If there are still some bugs left, you’ll spot them soon enough.
Refresh script
Achilles among others creates a achilles_result_concept_count table. However, Achilles does only create a record_count, and not a person_count, which you’ll need for some Atlas functionality. Therefore, we need to separately refresh this table using this query provided by the WebAPI.
Too bad this query is written in PostgreSQL. However, we’ve made an attempt to translate it.
/************************************************/
/***** Create record and person count table *****/
/************************************************/
DROP TABLE IF EXISTS results.achilles_result_concept_count;
CREATE TABLE results.achilles_result_concept_count
(
concept_id int,
record_count bigint,
descendant_record_count bigint,
person_count bigint,
descendant_person_count bigint
);
/**********************************************/
/***** Populate record/person count table *****/
/**********************************************/
INSERT INTO results.achilles_result_concept_count (concept_id, record_count, descendant_record_count, person_count, descendant_person_count)
WITH counts AS (
SELECT stratum_1 concept_id, MAX (count_value) agg_count_value
FROM results.achilles_results
WHERE analysis_id IN (2, 4, 5, 201, 225, 301, 325, 401, 425, 501, 505, 525, 601, 625, 701, 725, 801, 825,
826, 827, 901, 1001, 1201, 1203, 1425, 1801, 1825, 1826, 1827, 2101, 2125, 2301)
/* analyses:
Number of persons by gender
Number of persons by race
Number of persons by ethnicity
Number of visit occurrence records, by visit_concept_id
Number of visit_occurrence records, by visit_source_concept_id
Number of providers by specialty concept_id
Number of provider records, by specialty_source_concept_id
Number of condition occurrence records, by condition_concept_id
Number of condition_occurrence records, by condition_source_concept_id
Number of records of death, by cause_concept_id
Number of death records, by death_type_concept_id
Number of death records, by cause_source_concept_id
Number of procedure occurrence records, by procedure_concept_id
Number of procedure_occurrence records, by procedure_source_concept_id
Number of drug exposure records, by drug_concept_id
Number of drug_exposure records, by drug_source_concept_id
Number of observation occurrence records, by observation_concept_id
Number of observation records, by observation_source_concept_id
Number of observation records, by value_as_concept_id
Number of observation records, by unit_concept_id
Number of drug era records, by drug_concept_id
Number of condition era records, by condition_concept_id
Number of visits by place of service
Number of visit_occurrence records, by discharge_to_concept_id
Number of payer_plan_period records, by payer_source_concept_id
Number of measurement occurrence records, by observation_concept_id
Number of measurement records, by measurement_source_concept_id
Number of measurement records, by value_as_concept_id
Number of measurement records, by unit_concept_id
Number of device exposure records, by device_concept_id
Number of device_exposure records, by device_source_concept_id
Number of location records, by region_concept_id
*/
GROUP BY stratum_1
UNION ALL
SELECT stratum_2 concept_id, SUM (count_value) AS agg_count_value
FROM results.achilles_results
WHERE analysis_id IN (405, 605, 705, 805, 807, 1805, 1807, 2105)
/* analyses:
Number of condition occurrence records, by condition_concept_id by condition_type_concept_id
Number of procedure occurrence records, by procedure_concept_id by procedure_type_concept_id
Number of drug exposure records, by drug_concept_id by drug_type_concept_id
Number of observation occurrence records, by observation_concept_id by observation_type_concept_id
Number of observation occurrence records, by observation_concept_id and unit_concept_id
Number of observation occurrence records, by measurement_concept_id by measurement_type_concept_id
Number of measurement occurrence records, by measurement_concept_id and unit_concept_id
Number of device exposure records, by device_concept_id by device_type_concept_id
but this subquery only gets the type or unit concept_ids, i.e., stratum_2
*/
GROUP BY stratum_2
), counts_person AS (
SELECT stratum_1 as concept_id, MAX (count_value) agg_count_value
FROM results.achilles_results
WHERE analysis_id IN (200, 240, 400, 440, 540, 600, 640, 700, 740, 800, 840, 900, 1000, 1300, 1340, 1800, 1840, 2100, 2140, 2200)
/* analyses:
Number of persons with at least one visit occurrence, by visit_concept_id
Number of persons with at least one visit occurrence, by visit_source_concept_id
Number of persons with at least one condition occurrence, by condition_concept_id
Number of persons with at least one condition occurrence, by condition_source_concept_id
Number of persons with death, by cause_source_concept_id
Number of persons with at least one procedure occurrence, by procedure_concept_id
Number of persons with at least one procedure occurrence, by procedure_source_concept_id
Number of persons with at least one drug exposure, by drug_concept_id
Number of persons with at least one drug exposure, by drug_source_concept_id
Number of persons with at least one observation occurrence, by observation_concept_id
Number of persons with at least one observation occurrence, by observation_source_concept_id
Number of persons with at least one drug era, by drug_concept_id
Number of persons with at least one condition era, by condition_concept_id
Number of persons with at least one visit detail, by visit_detail_concept_id
Number of persons with at least one visit detail, by visit_detail_source_concept_id
Number of persons with at least one measurement occurrence, by measurement_concept_id
Number of persons with at least one measurement occurrence, by measurement_source_concept_id
Number of persons with at least one device exposure, by device_concept_id
Number of persons with at least one device exposure, by device_source_concept_id
Number of persons with at least one note by note_type_concept_id
*/
GROUP BY stratum_1
), concepts AS (
select concept_id as ancestor_id, coalesce(cast(ca.descendant_concept_id as STRING), concept_id) as descendant_id
from (
select concept_id from counts
UNION DISTINCT
-- include any ancestor concept that has a descendant in counts
select distinct cast(ancestor_concept_id as STRING) concept_id
from counts c
join cdm_pulse_01.concept_ancestor ca on cast(ca.descendant_concept_id as STRING) = c.concept_id
) c
left join cdm_pulse_01.concept_ancestor ca on c.concept_id = cast(ca.ancestor_concept_id as STRING)
)
SELECT
cast(concepts.ancestor_id as int) concept_id,
coalesce(max(c1.agg_count_value), 0) record_count,
coalesce(sum(c2.agg_count_value), 0) descendant_record_count,
coalesce(max(c3.agg_count_value), 0) person_count,
coalesce(sum(c4.agg_count_value), 0) descendant_person_count
FROM concepts
LEFT JOIN counts c1 ON concepts.ancestor_id = c1.concept_id
LEFT JOIN counts c2 ON concepts.descendant_id = c2.concept_id
LEFT JOIN counts_person c3 ON concepts.ancestor_id = c3.concept_id
LEFT JOIN counts_person c4 ON concepts.descendant_id = c4.concept_id
GROUP BY concepts.ancestor_id;Cache
No, we’re not done yet! There’s still cache. Atlas does not retrieve the record counts directly from achilles_result_concept_count, but uses a caching table. To update the record counts, we need to truncate the caching table, after which it will be automatically filled with the new record counts. The cache resides in the webapi.cdm_cache table in the broadsea-atlasdb container.
sudo docker exec -it broadsea-atlasdb /bin/bash
psql --host localhost --username postgres
truncate table webapi.cdm_cache;Debugging
When everything works at once, it’s great. Usually it doesn’t. Atlas depends greatly on the WebAPI. When something isn’t working as it is supposed to, the WebAPI logs are typically a good place to start searching.
sudo docker logs ohdsi-webapiNotes
Feel free to check our forks of the Broadsea and WebAPI repositories:
Also, feel free to reach out via mail if you have any questions: contact details